CCI Zegami Server
Below are a list of datasets hosted on the the Liverpool CCI Zegami server. If you are interested in hosting your data for private or public use, please contact a member of staff: cREMOVEMEcREMOVEMEi@REMOVEMEliv.aREMOVEMEc.REMOVEMEuk.
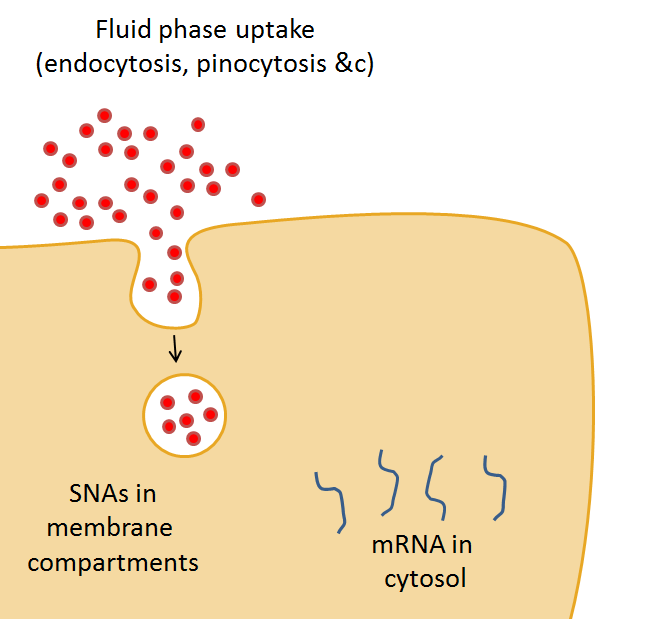
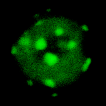
SmartFlares
This dataset supports a 2016 Open Science project from the Lévy lab investigating the ability of Spherical Nucleic Acids (SmartFlares) to gain access to the cell cytoplasm.
The project was published in March 2016 (DOI:10.14293/S2199-1006.1.SOR-CHEM.AZ1MJU.v2). More information can be found in the Open Science notebook, or via this blog post.
David Mason (Léy Lab) - March 2016

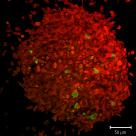
Spheroid Tracking Data
This dataset shows the results of post-acquisition processing of 4D tracking data in a spheroid invasion system. Spheroids were grown using H2B-mRFP labelled cells and imaged using lightsheet microscopy. The cells in the spheroids were tracked using Imaris and the track data post-processed using MATLAB to create these descriptive graphs. Maximum Intensity Projections are also shown for t=0,8,16 and 24h.
Rosalie Richards (Sée Lab) and David Mason (CCI)
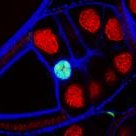
Studying Drosophila Development
Various genetic factors control tissue size control in developing Drosophila wing disks. Here, some of these genes are overexpressed and the resulting phenotypes were characterised within the context of growth and development.
Vincent Jonchere (Bennett Lab) - August 2016
There are some great example datasets that include OMERO integration and geographic mapping on the Zegami website.